
lm can be used to fit a two-way ANOVA model: twoway.model |t|) Suppose our experiment involves two factors, treatment and time. # 1.1724940 0.4455249 1.9153967 4.2413688 9.1016661 -1.6877019įor a model with more than one coefficient, summary provides estimates and tests for each coefficient adjusted for all the other coefficients in the model. # F-statistic: 12.49 on 5 and 19 DF, p-value: 1.835e-05 coef(batch.model) # (Intercept) treatmentB treatmentC treatmentD treatmentE batchBatch2 # Residual standard error: 1.735 on 19 degrees of freedom We do this by adding the covariate “batch” to the model formula: batch.model |t|) Suppose we want to adjust for batch differences in our model.

Treatment B - treatment A, no-intercept model: coefs <- coef(no.intercept.model)Ĭoefs - coefs # treatmentBįor the RNASeq analysis programs limma and edgeR, the model is specified through the design matrix. Treatment B - treatment A, reference group coded model: coefs <- coef(oneway.model) The no-intercept model is the SAME model as the reference group coded model, in the sense that it gives the same estimate for any comparison between groups: Without the intercept, the coefficients here estimate the mean in each level of treatment: treatmentmeans # A B C D E # F-statistic: 31.66 on 5 and 20 DF, p-value: 7.605e-09 coef(no.intercept.model) # treatmentA treatmentB treatmentC treatmentD treatmentE
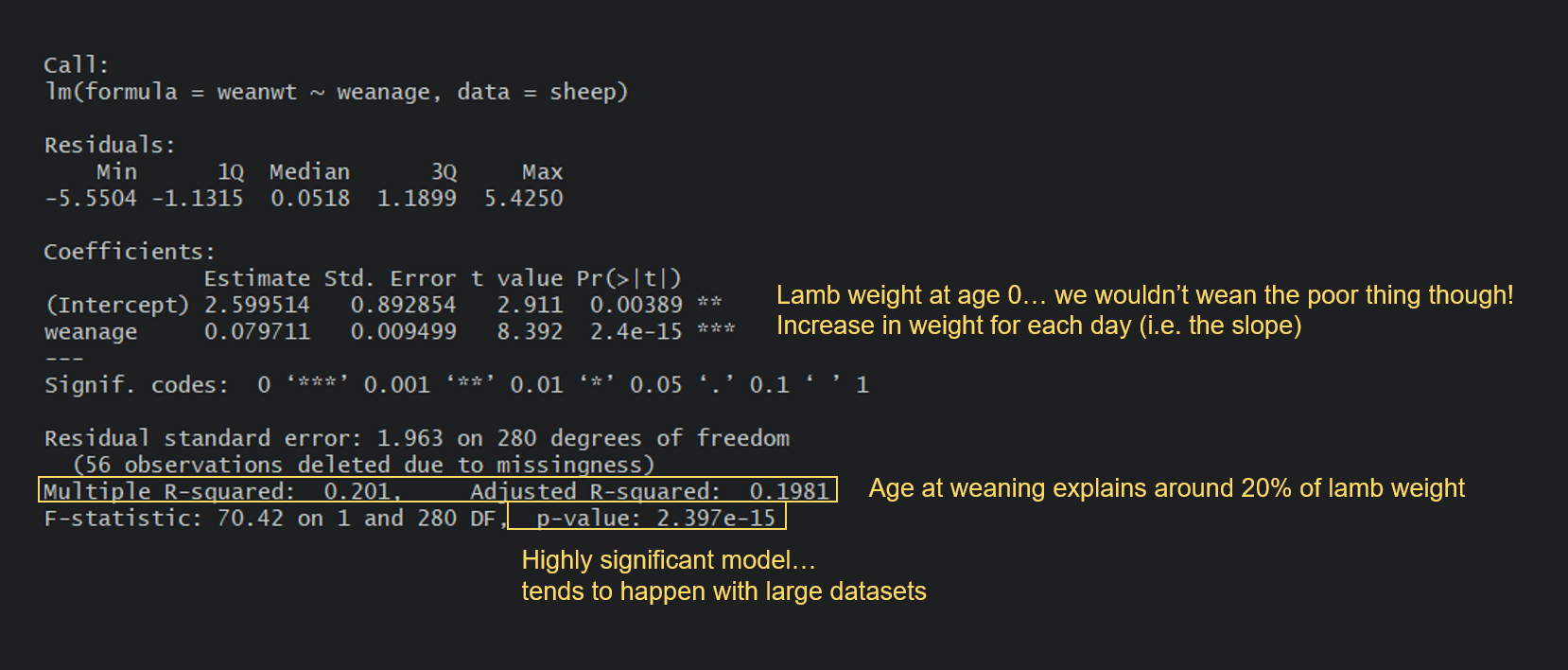
The residual standard error is the estimate of the variance of \(\epsilon\).“Pr(>|t|)” is the p-value for the coefficient.“t value” is the coefficient divided by its standard error.Error” is the standard error of the estimate

“Estimate” is the estimate of each coefficient.“Coefficients” refer to the \(\beta\)’s.
# Residual standard error: 1.74 on 20 degrees of freedom R uses the function lm to fit linear models.


 0 kommentar(er)
0 kommentar(er)
